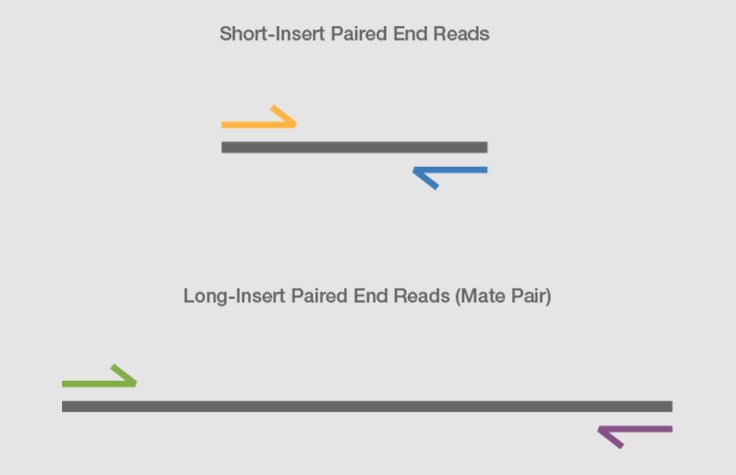
paired end sequencing read length
An analysis by Whiteford et al. My paired-end read sequencing data are 50 base-pairs on each end nearly all exact with about 300bp unsequenced inner distance between the 50bp ends.
There are commercial sequencing kitssequencers that allow for paired end sequencing in which the two reads obtained for each fragment are of different length.

. The term paired ends refers to the two ends of the same DNA molecule. To ensure sequencing quality of the Index Read do not exceed the supported read. The library prep protocols are designed to.
Paired-end reads are required to get information from both 5 and 3 ends of RNA species with stranded RNA-Seq library preparation kits. On sequencing using unpaired reads shows that ultra-short reads theoretically allow whole genome re-sequencing and de novo assembly of only small. Small RNA Analysis Due to the short.
To ensure sequencing quality of the Index Read do not exceed the supported read. Application of sequencing to RNA analysis RNA-Seq whole transcriptome SAGE expression analysis novel organism mining splice variants Search in titles only Search in RNA. During sequencing it is possible to specify the number of base pairs that are read at a time.
However some paired-end sequencing data show the presence of a subpopulation of reads where the second read R2 has lower average qualities. The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. Mate pairs involves making circular.
For example one read might consist of 50 base pairs 100 base pairs or more. Simple workflow allows generation of unique ranges of insert sizes. Paired ends is supported by some technologies Illumina and Sanger where it is possible to sequence from both ends of a clone.
The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. Requires the same amount. This size depends on the library.
A good choice for read length is closely tied to the insert size of the sequencing library ie how long the individual DNA fragments are that are sequenced. Reading the nucleotides from two ends of a DNA fragment is called paired-end tag PET sequencing. We use an Illumina MiniSeq for our short-read sequencing runs.
The paired-end short read lengths are always 2 x 150bp 300bp. Here are my answers. We show that the fragment.
Enter up to 20 characters or use manual mode if you need between 20 and 100 bp. Enter up to 20 characters or use manual mode if you need between 20 and 100 bp. 08-20-2008 0508 PM.
When the fragment length is longer than the combined read length. So you can sequence one end then turn it around and sequence the other end.
Frontiers Long Read Sequencing Emerging In Medical Genetics

Skewer A Fast And Accurate Adapter Trimmer For Next Generation Sequencing Paired End Reads Topic Of Research Paper In Biological Sciences Download Scholarly Article Pdf And Read For Free On Cyberleninka Open Science

Illumina Sequencing Georgia Genomics And Bioinformatics Core Ggbc
Illumina Hiseq 2500 And Miseq Ngs Core At Brcas
What Is Paired End 150 Omega Bioservices
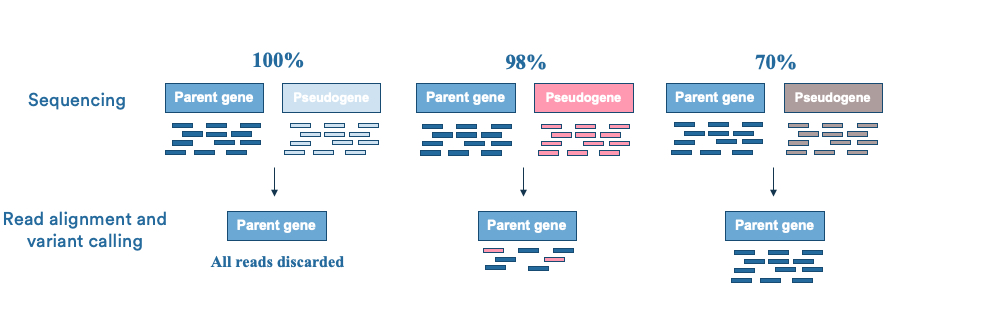
Blueprint Genetics Approach To Pseudogenes And Other Duplicated Genomic Regions Blueprint Genetics

Assessing The Performance Of The Oxford Nanopore Technologies Minion Sciencedirect

Pdf Local De Novo Assembly Of Rad Paired End Contigs Using Short Sequencing Reads Semantic Scholar

How To Calculate The Coverage For A Ngs Experiment
Local De Novo Assembly Of Rad Paired End Contigs Using Short Sequencing Reads Plos One
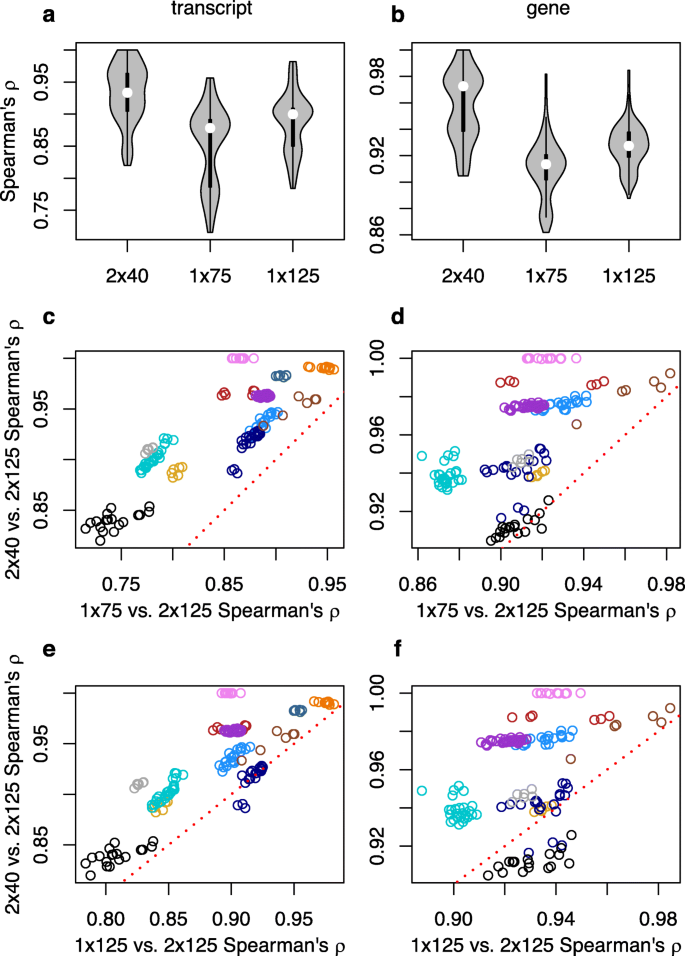
Short Paired End Reads Trump Long Single End Reads For Expression Analysis Bmc Bioinformatics Full Text
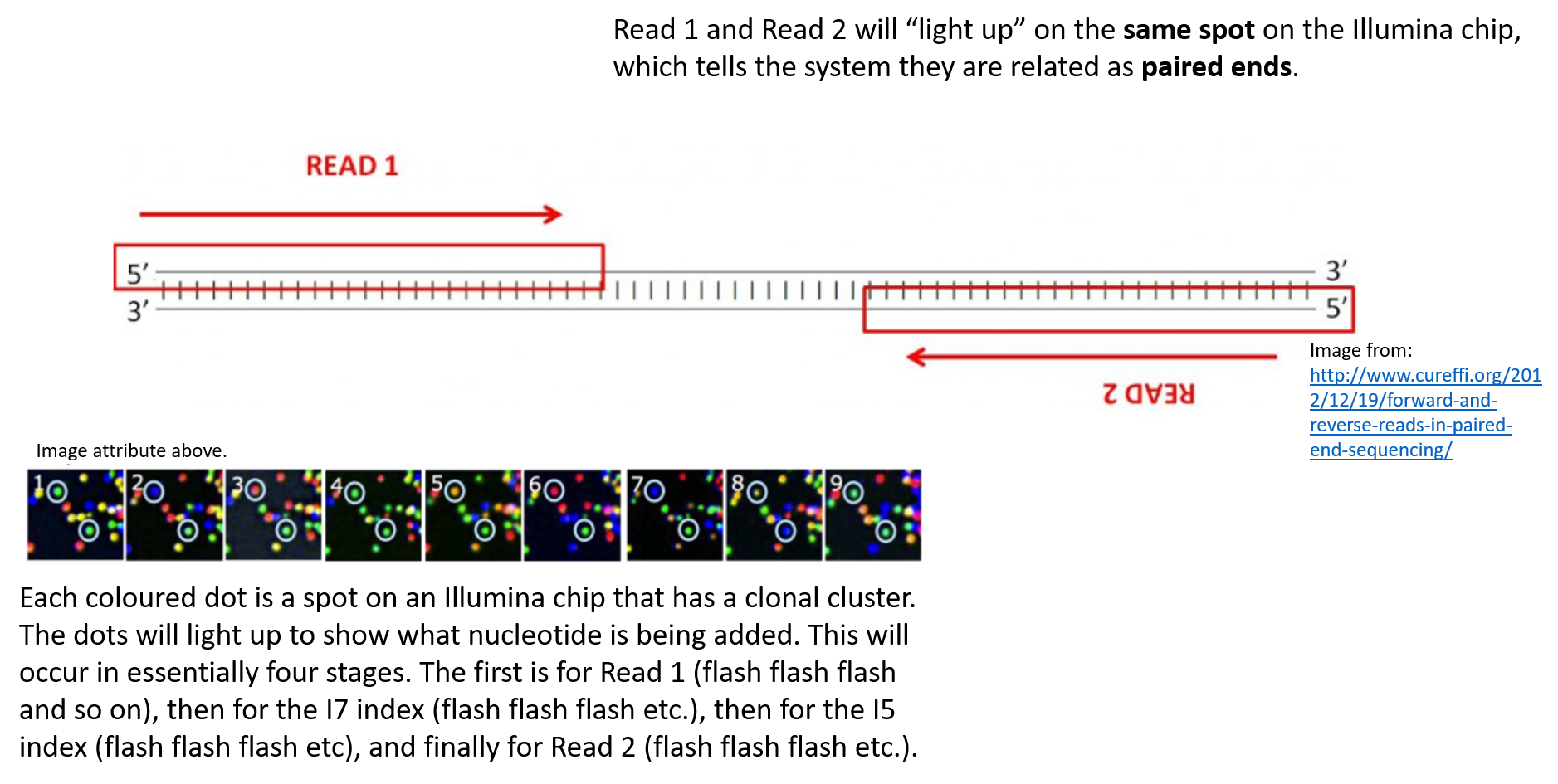
Illumina Sequencing For Dummies An Overview On How Our Samples Are Sequenced Kscbioinformatics

Casava Files Of Paired End Reads General Discussion Qiime 2 Forum